chameleon session help guide
Getting ready for your chameleon session
You have been assigned a chameleon session at NCCAT. What are the next steps?
Have a pre-session meeting with the NCCAT User Office and fill out the [pre-award questionnaire] before the meeting.
Make sure you have completed the pre-session forms:
[User registration][Shipping manifest][Shipping agreement]Review our shipping guide [here].
Verify the spokesperson of the proposal can login with your account at nccatweb.nysbc.org .
Register an account with Globus.org so you may download your data. See our quick guide or Globus’ FAQ.
The workflow for this access category is as follows:
- users will ship their sample to NCCAT,
- our operators will iteratively optimize the experiment to maximize the performance of chameleon, and
- we will send you back 1-2 grid boxes with up to four (4) grids in each grid box, and a feedback report.
If you would like NCCAT to collect data then we can hold onto your grids pending you have a Krios access proposal.
Sample considerations for your chameleon session
We make the following recommendations for chameleon:
- No less than 50 μl of protein sample.
- High purity, non-aggregated, stable samples.
- Minimal buffer additives (ideally no organic solvents or viscous reagents).
- Include all chemicals/ligands required for protein stability.
- No less than 2.5x the concentration used for plunge freezing. Typically on the order of several mg/mL for standard samples.
- Include 5 mL of dilution buffer that matches the sample storage buffer.
- Do not lyophilize your protein.
During our pre-session meeting make sure you have the following information:
- Name/Title of macromolecule of interest:
- Molecular weight:
- Storage buffer:
- Ligands/Binding partners in sample (if applicable):
- Standard storage temperature (in oC):
- Storage time (max time at storage temperature and/or at RT):
- Highest soluble concentration tested (mg/mL):
- Issue the sample is facing ( aagregation, preffered orientation, denaturation, etc.):
Your chameleon session
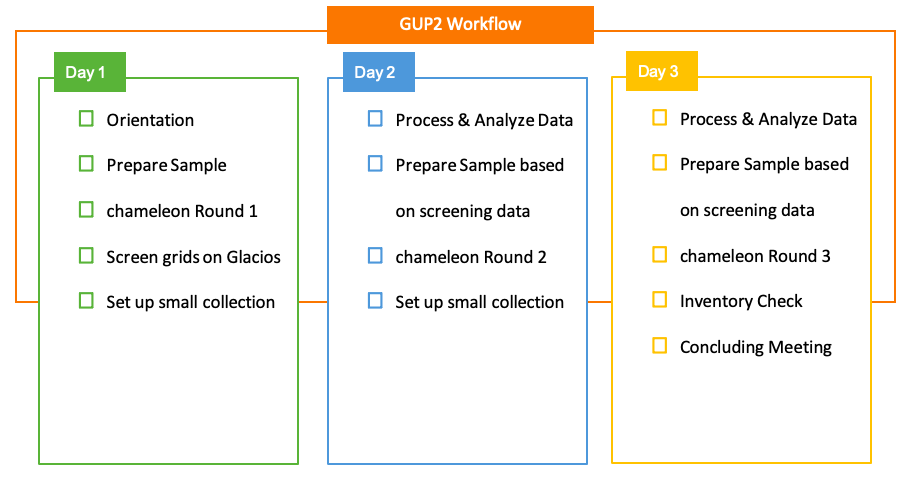
A representative schedule for a half-week chameleon session at NCCAT.
Leginon and Appion webtools
nccatweb.nysbc.org
After a Chameleon session, we give representative feedback on the samples using Leginon on an NCCAT microscope. For responsive feedback, Leginon has webtools so the user may monitor the data collection and be able to also analyze the quality of the grids made.
By default, we collect data under the spokesperson’s account. If additional accounts are required, these requests may be made to the NCCAT User Office during your pre-session meeting, or email correspondences, ahead of a scheduled data collection session.
If you have trouble with your accounts you may email the NCCAT User Office.
Image viewer
The main way to monitor your data collection is through the image viewer through our web interface.
Note that NCCAT’s business hours are 9am-5pm ET during business days. Email correspondences outside this time may be delayed.
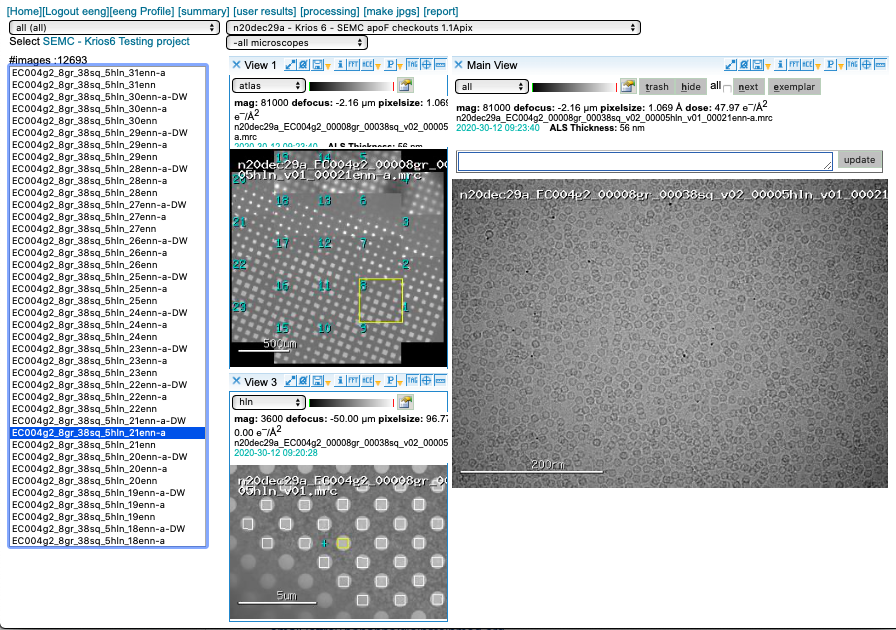
3-way viewer
Appion pre-processing
Appion pre-processing has a few useful features. Although we prioritize data collection (acquisition of movies) we strive to provide as much feedback as possible.
Motion Correction
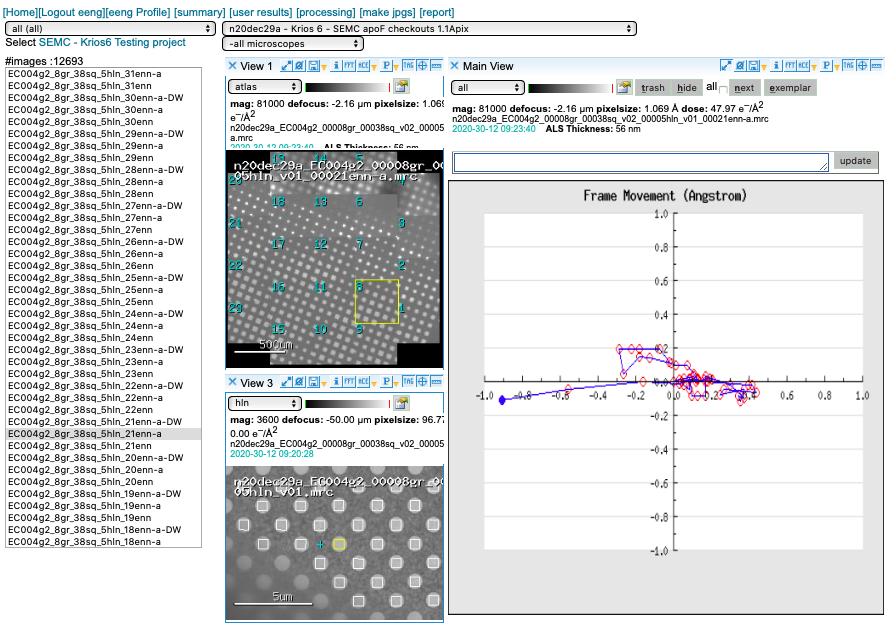
We run MotionCor2 using 5×5 patches. Bin1 for counting and Bin2 for super-res. You may access this plot through the arrow icon.
CTF estimation
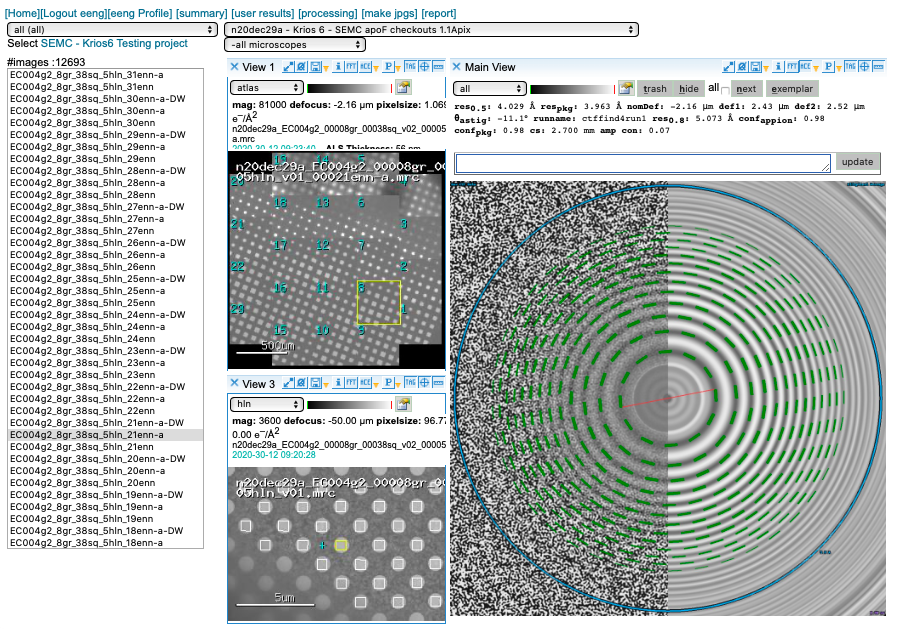
We prioritize our GPUs for motion correction and use CTFFind4 on our spare CPUs for CTF estimation. You may access this plot through the “ACE” icon.
Image tool
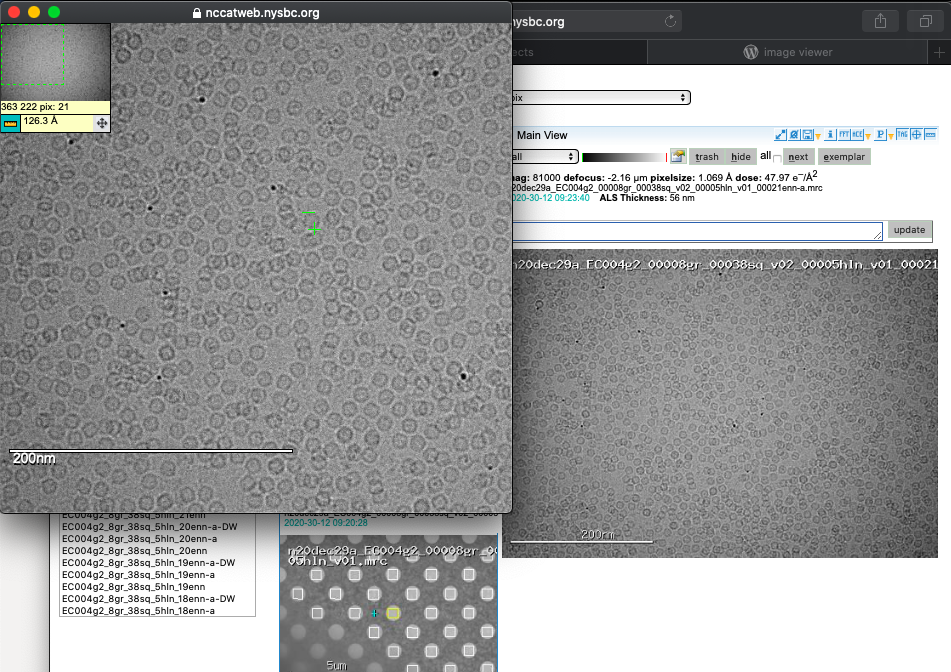
A common question is what size is my particle? By clicking on the image you can expand your view and engage a ruler tool to measure your macromolecules of interest.
Reports
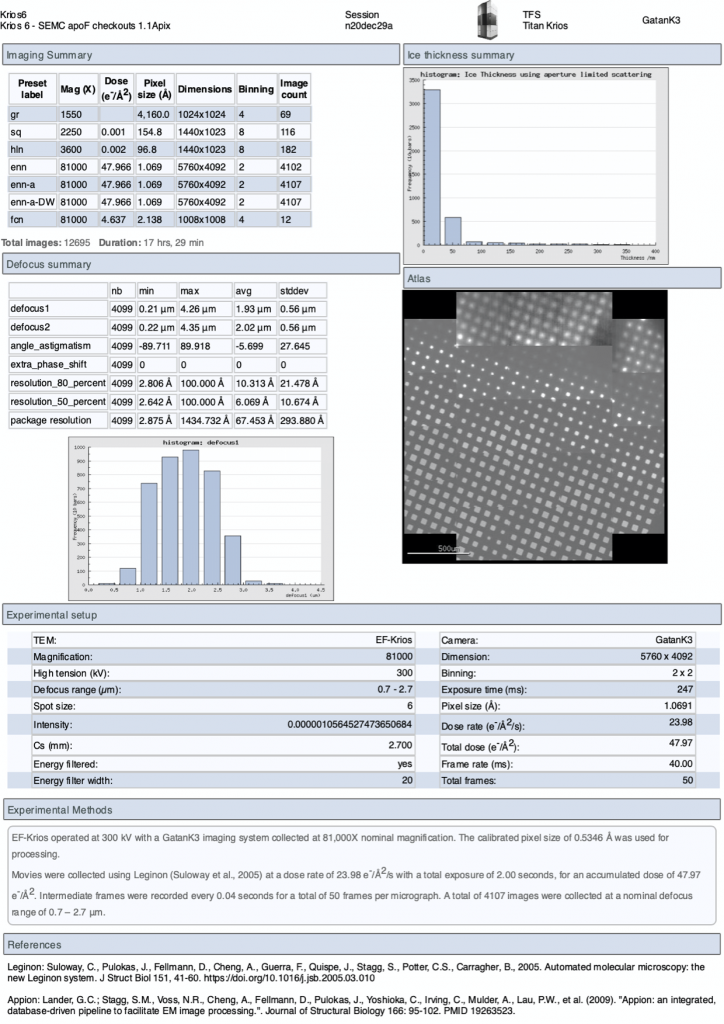
On the upper menu in the webpage you may also access a “Report” page to give you a summary of the experiment. This report updates in real-time.
What are all these presets?
- gr – images montaged together to create a whole grid Atlas
- sq – medium magnification image used to find squares
- hl – medium magnification image used to find holes
- enn or esn – high magnification image in counting or super res, respectively
The Leginon files names are explicit in case you lose web viewer functionality you can trackback to the parent images related to your current image.
Why are there 3 versions of enn?
Leginon aligned sum images are as follows:
- *enn.mrc – unaligned sums and may be 8×8 early return images
- *enn-a.mrc – aligned sums (by MotionCor2)
- *enn-a-DW.mrc – aligned dose weighted sums (by MotionCor2)
Additional tools in our web viewer include for each image the nominal defocus, estimated pixel size, total dose of the movie, timestamp when the image was taken, and ice thickness monitoring. If you are curious about our Appion pre-processing tools you may schedule a small group training session through the NCCAT User Office.
After your session / getting your data
Please download your data via Globus. It may be easier to download the raw data after a session is finished, but the Leginon aligned sum images (*enn-a-DW.mrc) are relatively small and that would be useful if you wanted to use them for initial processing feedback. We will store your data for 1 month, then it will be queued for deletion. If you have issues downloading your data please let us know.
Your data is only accessible through the spokesperson’s login located in a few places:
- the data on nccatweb.nysbc.org is located in /beegfs/leginon/[username]/ ,
- the direct detector data is located in /beegfs/frames/[username] ,
- the pre-processing is in the Appion folder and may be regenerated through processing and located in /beegfs/appion/[username] , and
- for Falcon data the movies are in mrc format and already dark and gain corrected. (For images collected in TIFF LZW the references are located in the folder called references in the frames directory. You may see a couple because we updated the gain after we started the session. So the latest one is the one to use.)
Data directories
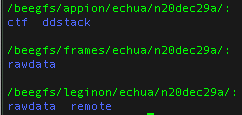
Note: At minimum users should always download all the data in the frames folder because this folder contains your raw data and all other files may be regenerated from your raw data.
References
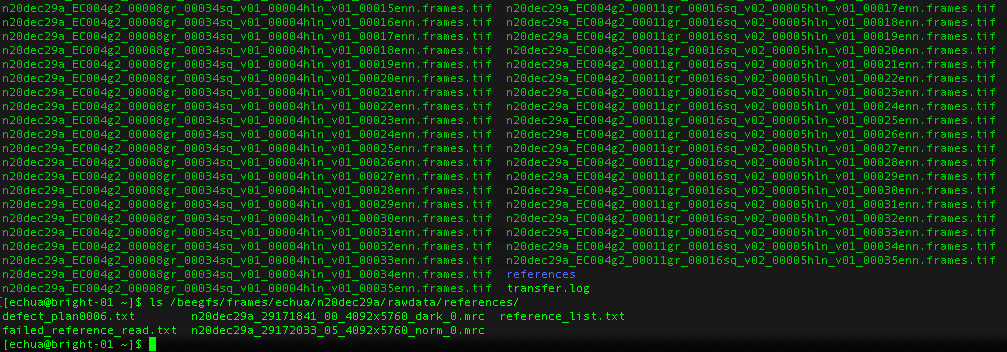
What about gain references?
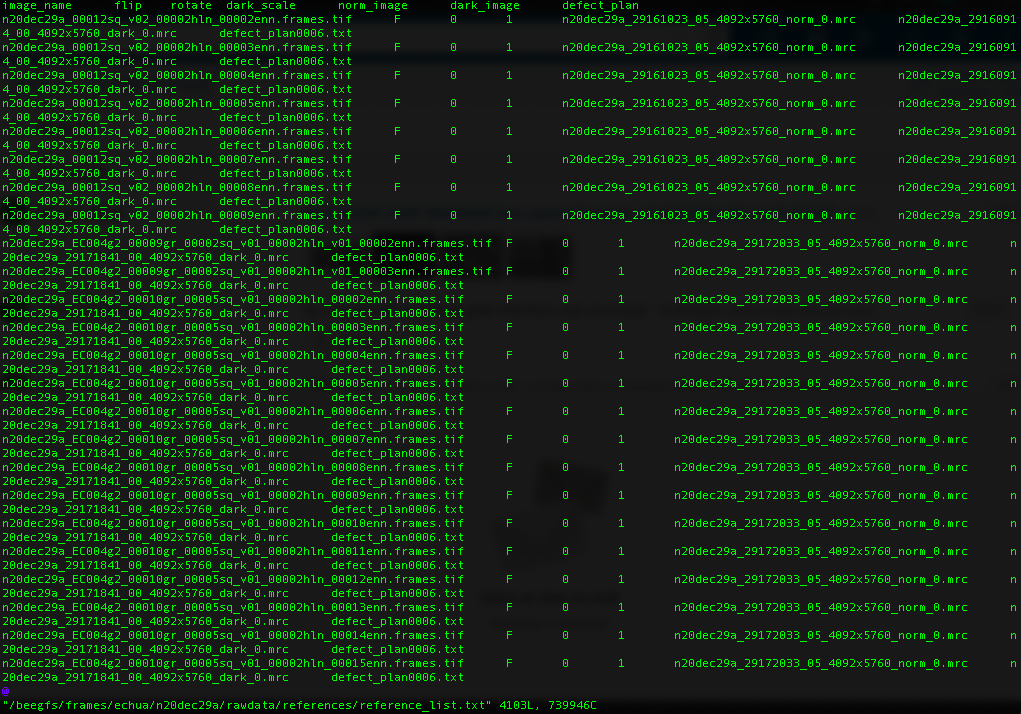
For Gatan K3 images gain correction requires a y-flip for most motion correction programs. The K3 is hardware dark corrected and only the gain reference is needed. The name of the gain reference would be in a format, such as “n20dec29a_29172033_05_4092x5760_norm_0.mrc”. If there are multiple references you should use the latest. If a new gain reference was taken in the middle of the session it would be annotated in the” reference_list.txt” file.
For Falcon III and Falcon IV data, there are no gain references required since the images are hardware gain and dark corrected.
For more information read our wiki [here].
Reference list for K3 data
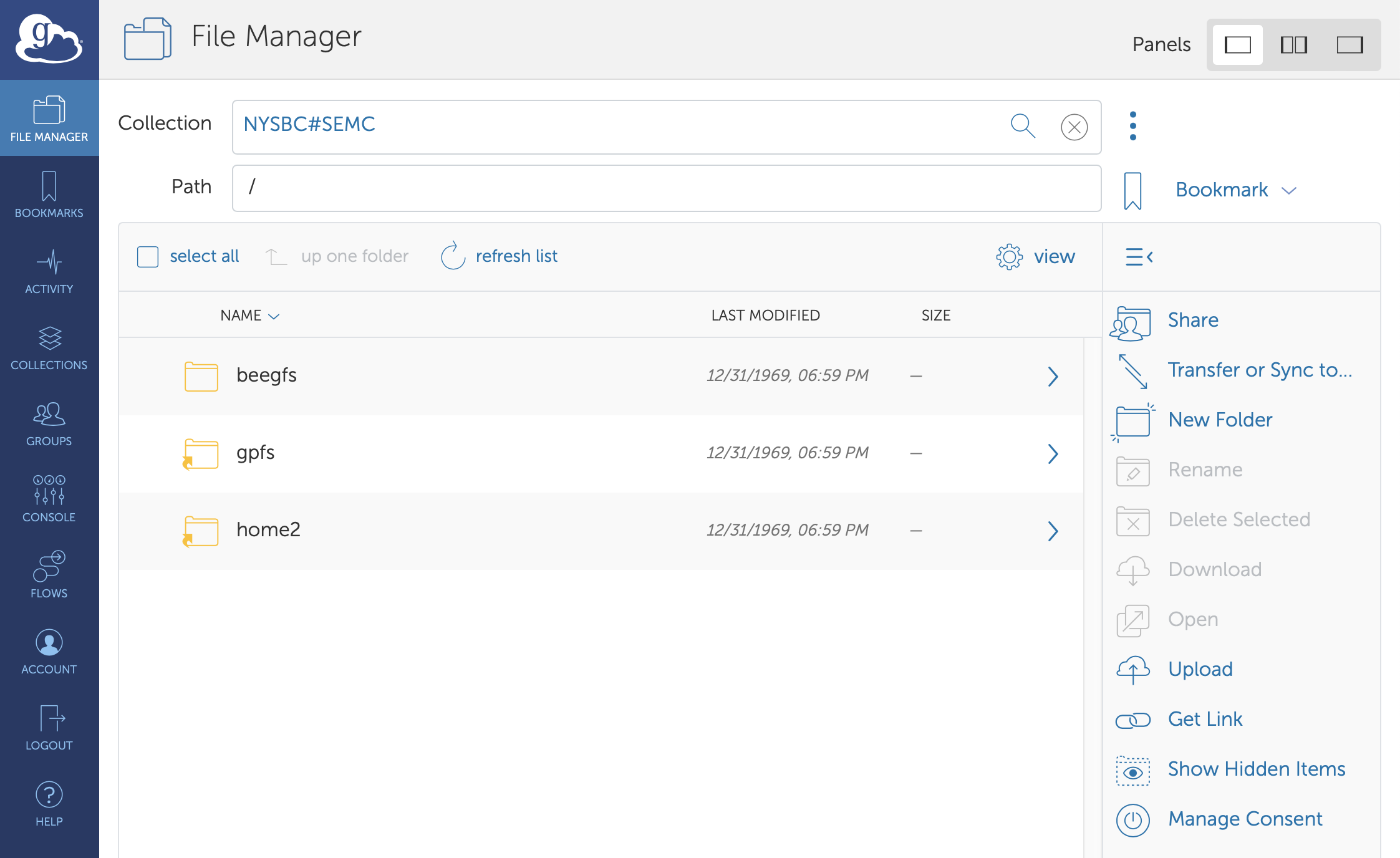
We use Globus, a non-profit service for secure, reliable research data management, for data transfer.
Please note that you are responsible for your data and samples. In general, direct detector data will be queued for deletion in a month. Your Leginon images are not backed up and may be converted to JPEG versions within 1 year. We will make our best efforts to preserve the functionality of the image viewer, but do not archive data. If you are having trouble with Globus let us know. If it is the endpoint we maintain our IT may assist, but Globus is a service we subscribe to and general help may also be found on their website: https://www.globus.org/
Globus file manager
At the end of your session
At the end of your session please make sure to contact the NCCAT User Office to work out any sample/grid logistics and fill out the end of session forms.
